Unraveling the Genetic Landscape: A Deep Dive into QTL Mapping
Related Articles: Unraveling the Genetic Landscape: A Deep Dive into QTL Mapping
Introduction
In this auspicious occasion, we are delighted to delve into the intriguing topic related to Unraveling the Genetic Landscape: A Deep Dive into QTL Mapping. Let’s weave interesting information and offer fresh perspectives to the readers.
Table of Content
Unraveling the Genetic Landscape: A Deep Dive into QTL Mapping

The study of genetics has revolutionized our understanding of life, revealing the intricate tapestry of traits passed down through generations. At the heart of this exploration lies the concept of Quantitative Trait Loci (QTL), genomic regions influencing complex traits that are not determined by a single gene but rather by the combined effects of multiple genes. Identifying these QTLs is crucial for understanding the genetic basis of traits like yield in crops, disease susceptibility in humans, and even behavior in animals.
Enter QTL mapping, a powerful tool that allows researchers to pinpoint these elusive genomic regions. This process involves analyzing the inheritance of traits alongside genetic markers, providing a blueprint of the genetic architecture underlying complex phenotypes.
The Fundamental Principles of QTL Mapping
QTL mapping rests on the principle of linkage disequilibrium, the non-random association of alleles at different loci. This association arises from the physical proximity of genes on chromosomes, meaning that genes located close together are more likely to be inherited together.
The process begins with the creation of a mapping population, typically a cross between two parental lines with contrasting traits. This population is then genotyped, meaning that the genetic markers are identified and mapped across the genome. Simultaneously, the phenotypes of interest are measured in the mapping population.
By comparing the inheritance of the traits with the genetic markers, researchers can identify regions of the genome where the trait variation is linked to marker variation. These regions are then declared QTLs, representing areas of the genome that contribute to the variation in the complex trait.
The Mechanics of QTL Mapping: A Step-by-Step Guide
-
Population Creation: The first step involves creating a mapping population. This is usually achieved through a cross between two contrasting parental lines, often referred to as the "donor" and "recipient" lines. These parental lines should exhibit clear differences in the trait of interest.
-
Phenotyping: The mapping population is then meticulously phenotyped, meaning that the trait of interest is measured in all individuals. This involves collecting data on the specific trait, which can be quantitative (e.g., height, yield, disease resistance) or qualitative (e.g., presence or absence of a trait).
-
Genotyping: The mapping population is then genotyped, which involves determining the genetic markers across the genome. This step utilizes various techniques, including microsatellite markers, single nucleotide polymorphisms (SNPs), and Restriction Fragment Length Polymorphisms (RFLPs).
-
QTL Analysis: The final step involves analyzing the data from the phenotyping and genotyping steps. Statistical methods are employed to identify regions of the genome where the variation in the trait is linked to marker variation. These regions are then declared QTLs.
Types of QTL Mapping Strategies
QTL mapping encompasses a range of strategies, each tailored to specific research objectives and population structures. Some common approaches include:
-
Interval Mapping: This approach analyzes the association between the trait and markers across the entire genome, identifying regions of linkage. It is a powerful tool for initial QTL detection.
-
Composite Interval Mapping: This method improves upon interval mapping by incorporating information from multiple markers simultaneously, enhancing the accuracy of QTL identification.
-
Multiple QTL Mapping: This approach seeks to identify multiple QTLs that contribute to the variation in the trait. It utilizes advanced statistical models to account for the complex interactions between QTLs.
Applications of QTL Mapping: Unveiling the Genetic Architecture of Complex Traits
QTL mapping has proven to be an indispensable tool in various fields, providing insights into the genetic basis of a wide range of complex traits. Some key applications include:
-
Crop Improvement: QTL mapping has revolutionized plant breeding by identifying genes controlling yield, disease resistance, and other desirable traits. This knowledge enables breeders to develop improved crop varieties with enhanced productivity and resilience.
-
Human Health: QTL mapping has contributed significantly to our understanding of human diseases, identifying genes associated with susceptibility to conditions like diabetes, heart disease, and cancer. This information is crucial for developing personalized medicine and targeted therapies.
-
Animal Breeding: QTL mapping has been instrumental in improving livestock breeding programs, identifying genes associated with traits like milk production, meat quality, and disease resistance. This has resulted in more efficient and sustainable animal production systems.
FAQs: Addressing Common Questions about QTL Mapping
1. What is the difference between QTL mapping and gene mapping?
While both involve identifying genetic regions, QTL mapping focuses on complex traits influenced by multiple genes, while gene mapping focuses on individual genes and their specific functions.
2. How many markers are needed for QTL mapping?
The number of markers required depends on the size of the genome, the population structure, and the desired resolution. A denser marker map generally provides more accurate QTL mapping.
3. What are the limitations of QTL mapping?
QTL mapping is susceptible to factors like population size, marker density, and the complexity of the trait being studied. It may not always pinpoint the exact gene responsible for the trait, but rather a region containing multiple genes.
4. How can QTL mapping be used for gene cloning?
QTL mapping can provide a starting point for gene cloning by identifying candidate regions. Further investigation, such as fine mapping and positional cloning, is then required to isolate the specific gene.
Tips for Effective QTL Mapping
-
Choose a mapping population with sufficient genetic diversity. This ensures a wider range of genetic variation for QTL detection.
-
Utilize a dense marker map. A higher density of markers improves the accuracy and resolution of QTL mapping.
-
Control environmental factors. Environmental variation can influence trait expression, potentially confounding QTL analysis.
-
Employ appropriate statistical methods. Selecting the correct statistical models is crucial for accurate QTL identification.
Conclusion: A Powerful Tool for Unveiling the Genetic Landscape
QTL mapping has emerged as a cornerstone of modern genetics, providing an invaluable tool for understanding the genetic architecture of complex traits. By identifying regions of the genome associated with trait variation, QTL mapping has revolutionized research in various fields, from agriculture to human health. As our understanding of genetics continues to evolve, QTL mapping will remain a vital tool for deciphering the complexities of life and harnessing the power of genetic information for the benefit of society.



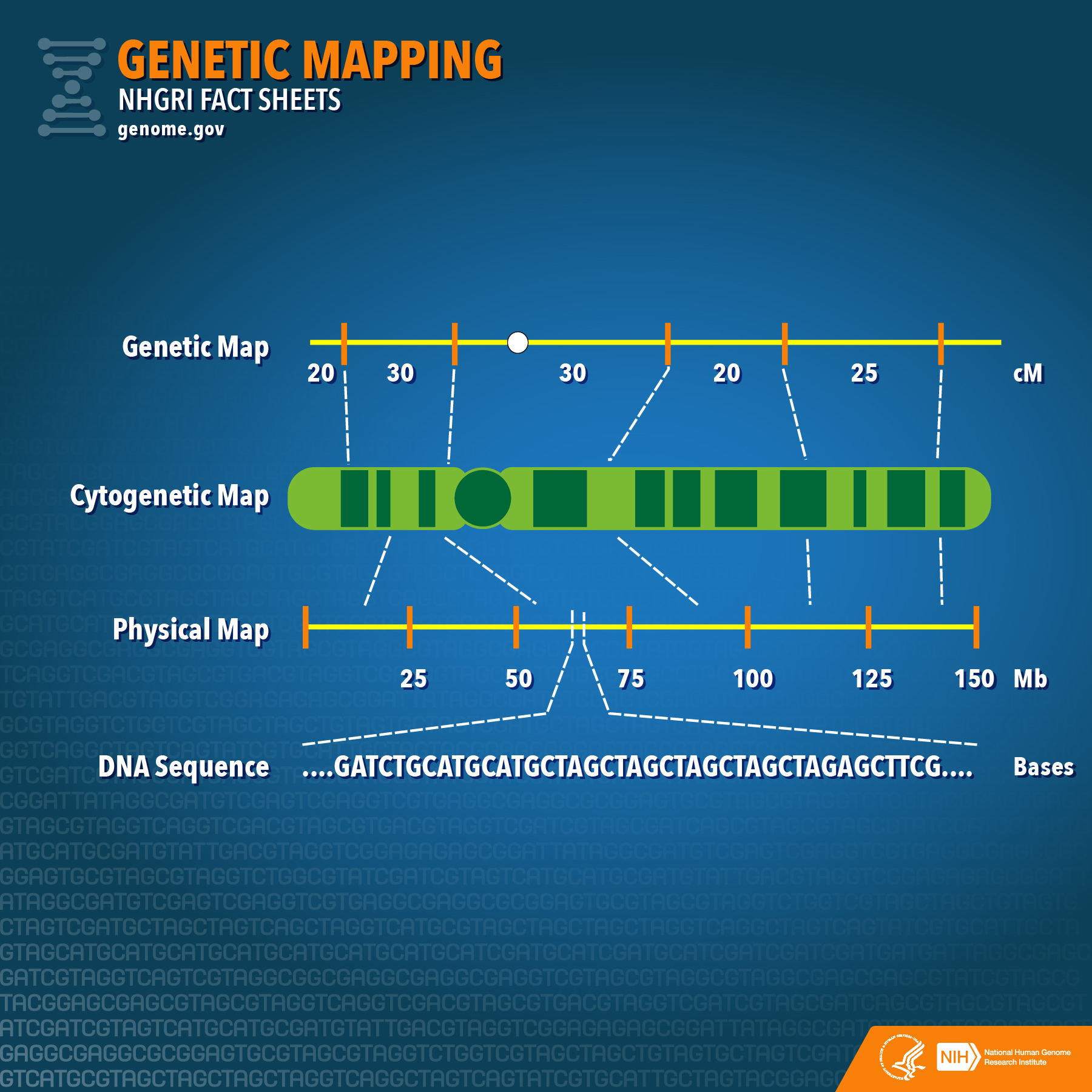

Closure
Thus, we hope this article has provided valuable insights into Unraveling the Genetic Landscape: A Deep Dive into QTL Mapping. We hope you find this article informative and beneficial. See you in our next article!